Selected Publications
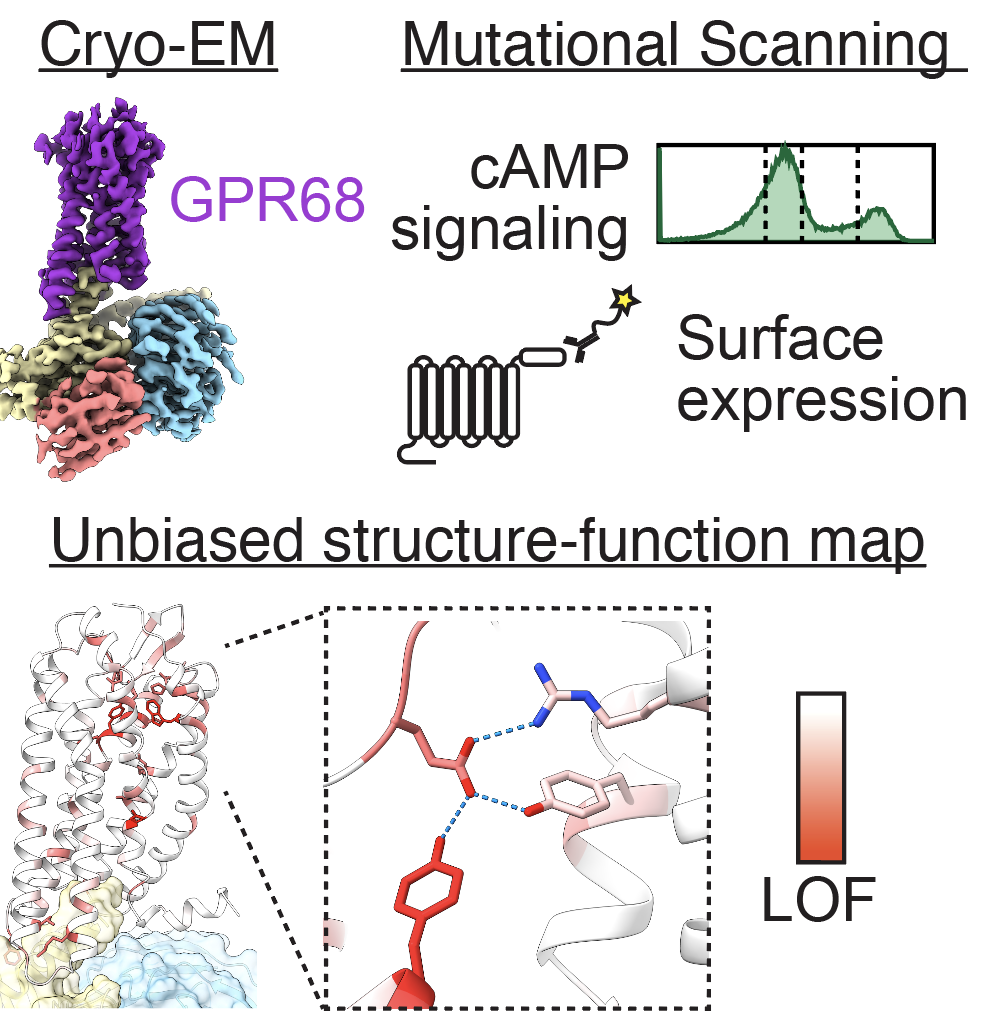
Molecular basis of proton-sensing by G protein-coupled receptors
Matthew K. Howard*, Nicholas Hoppe*, Xi-Ping Huang, Darko Mitrovic, Christian B. Billesbølle, Christian B. Macdonald, Eshan Mehrota, Patrick Rockefeller Grimes, Adam Zahm, Donovan D. Trinidad, Lucie Delemotte, Justin English, Willow Coyote-Maestas†, Aashish Manglik†
Cell,
2025
The full spectrum of OCT1 (SLC22A1) mutations bridges transporter biophysics to drug pharmacogenomics
Sook Wah Yee*, Christian Macdonald*, Darko Mitrovic*, Xujia Zhou, Megan L. Koleske, Jia Yang, Dina Buitrago Silva, Patrick Rockefeller Grimes, Donovan Trinidad, Swati S. More, Linda Kachuri, John S. Witte, Lucie Delemotte†, Kathleen M. Giacomini†, Willow Coyote-Maestas†
Molecular Cell,
2024
Deep insertion, deletion, and missense mutation libraries for exploring protein variation in evolution, disease, and biology
Macdonald CB, Nedrud D, Rockefeller Grimes P, Trinidad D, Fraser JS, Coyote-Maestas W
Genome Biology,
2023
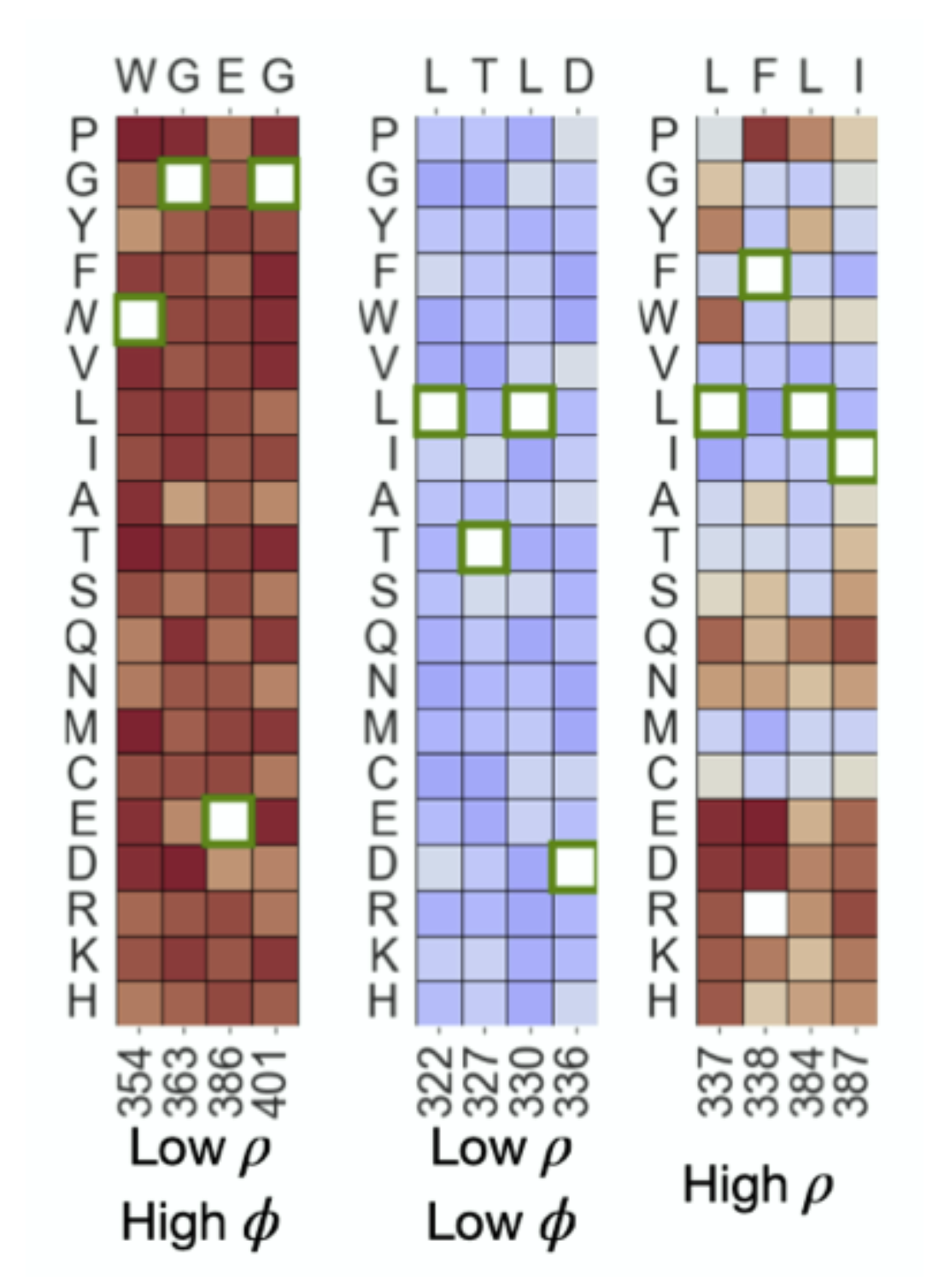
Determinants of trafficking, conduction, and disease within a K+ channel revealed through multiparametric deep mutational scanning
Willow Coyote-Maestas, David Nedrud, Yungui He, Daniel Schmidt
Elife,
2022
All Other Publications
Rosace-AA: Enhancing Interpretation of Deep Mutational Scanning Data with Amino Acid Substitution and Position-Specific Insights
Jingyou Rao, Mingsen Wang, Matthew Howard, Christian Macdonald, James S. Fraser, Willow Coyote-Maestas, Harold Pimentel
Biorxiv,
2025
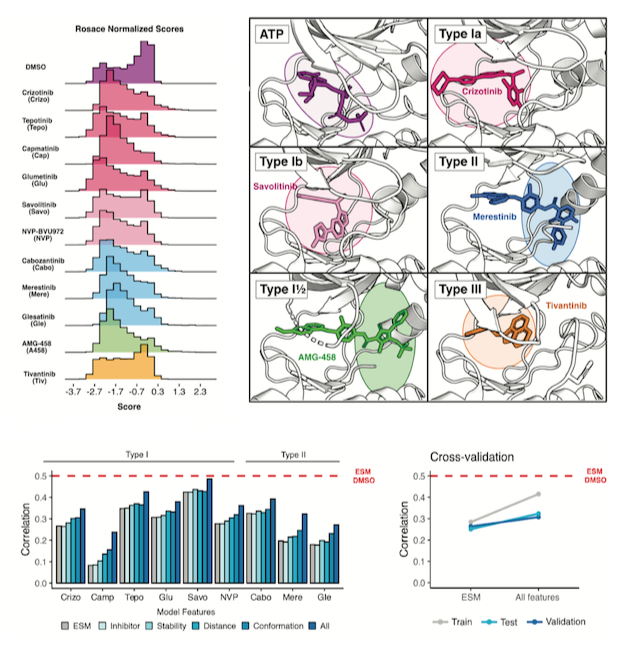
Mapping kinase domain resistance mechanisms for the MET receptor tyrosine kinase via deep mutational scanning
Estevam GO, Linossi EM, Jingyou Rao, Macdonald CM, Ravikumar A, Chrispens KM, Capra JA, Coyote-Maestas W, Pimentel H, Collisson EA, Jura N, Fraser JS
Biorxiv,
2024
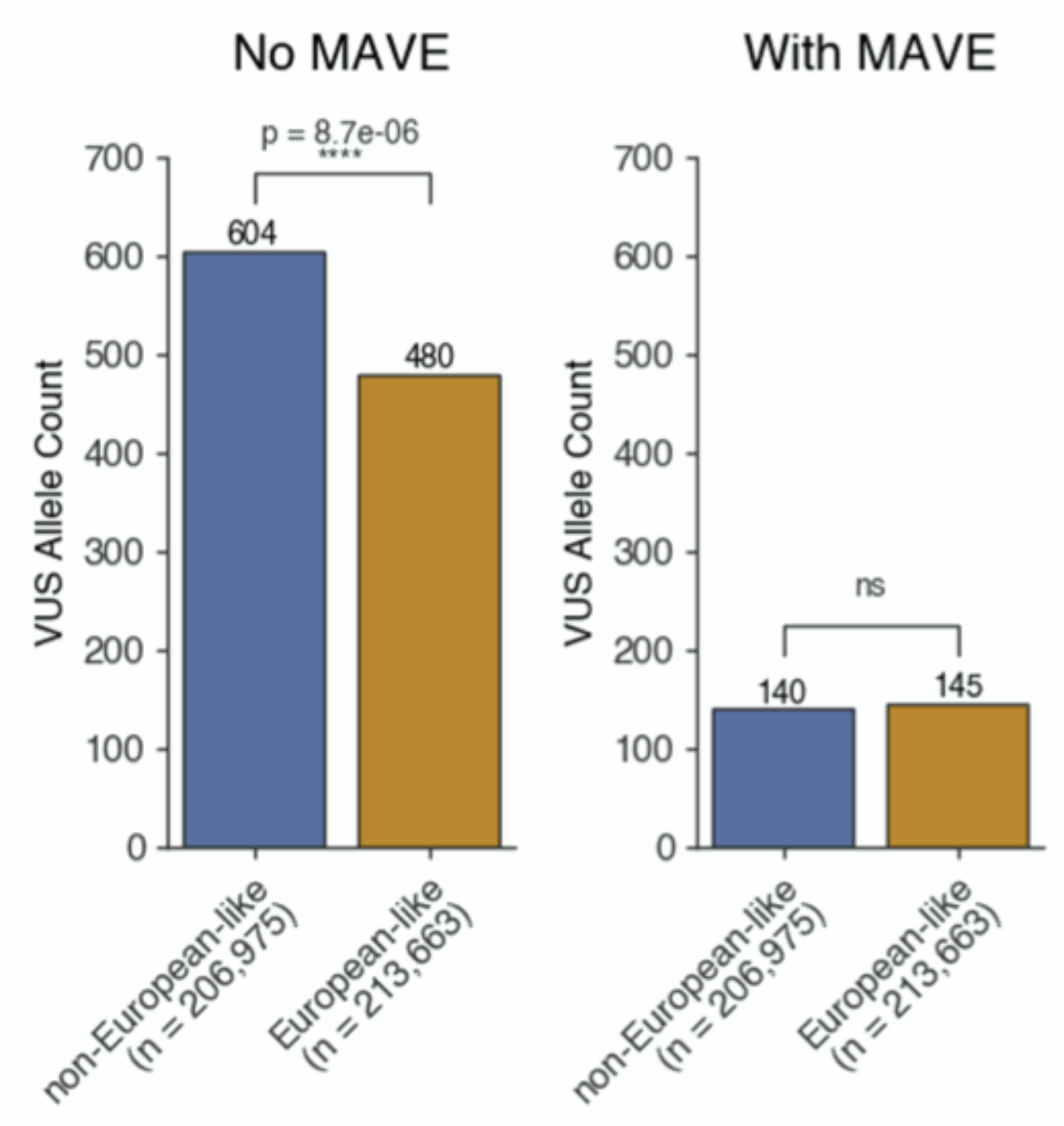
Using multiplexed functional data to reduce variant classification inequities in underrepresented populations
Moez Dawood†, Shawn Fayer, Sriram Pendyala, Mason Post, Divya Kalra, Karynne Patterson, Eric Venner, Lara A. Muffley, Douglas M. Fowler, Alan F. Rubin, Jennifer E. Posey, Sharon E. Plon, James R. Lupski, Richard A. Gibbs, Lea M. Starita, Carla Daniela Robles-Espinoza, Willow Coyote-Maestas†, Irene Gallego Romero†
Genome Medicine,
2024
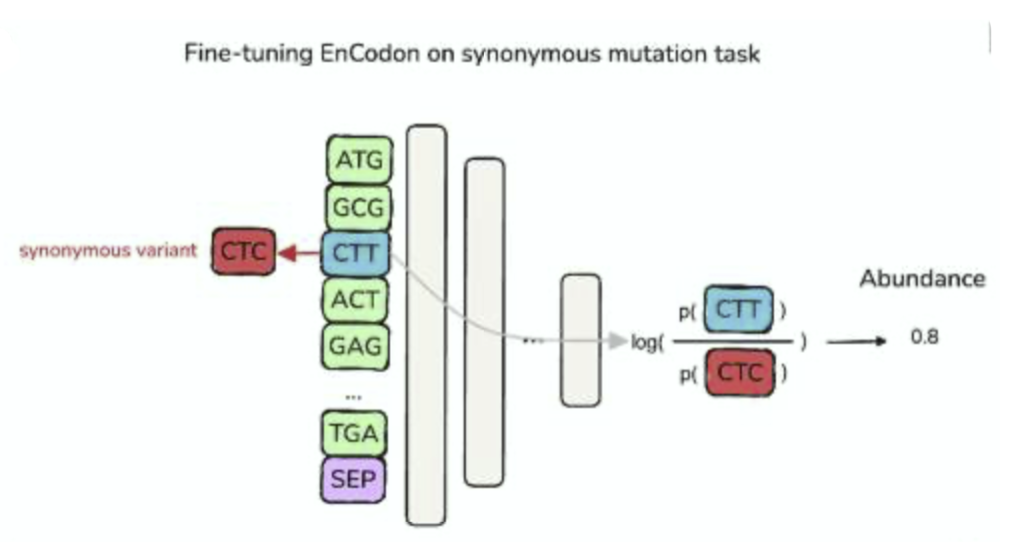
A Suite of Foundation Models Captures the Contextual Interplay Between Codons
Mohsen Naghipourfar, Siyu Chen, Mathew K Howard, Christian B Macdonald, Ali Saberi, Timo Hagen, Mohammad R K Mofrad†, Willow Coyote-Maestas†, Hani Goodarzi†
Biorxiv,
2024
Conserved regulatory motifs in the juxtamembrane domain and kinase N-lobe revealed through deep mutational scanning of the MET receptor tyrosine kinase domain
Estevam GO, Linossi EM, Macdonald CM, Espinoza CA, Michaud JM, Coyote-Maestas W, Collisson EA, Jura N, Fraser JS
eLife,
2024
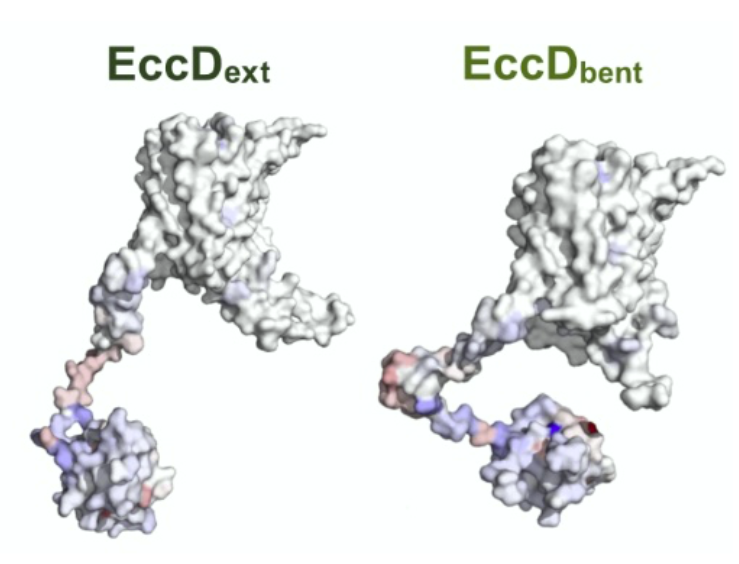
Deep mutational scanning of EccD3 reveals the molecular basis of its essentiality in the mycobacterium ESX secretion system
Donovan D Trinidad, Christian B Macdonald, Oren S Rosenberg, James S Fraser, Willow Coyote-Maestas
Biorxiv,
2024
Rosace: a robust deep mutational scanning analysis framework employing position and mean-variance shrinkage
Jingyou Rao, Ruiqi Xin*, Christian Macdonald*, Matthew Howard, Gabriella O. Estevam, Sook Wah Yee, Mingsen Wang, James S. Fraser, Willow Coyote-Maestas†, Harold Pimentel†
Genome Biology,
2024
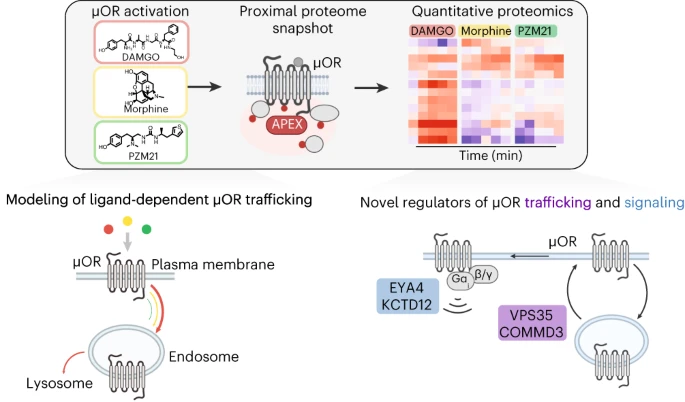
Profiling the proximal proteome of the activated μ-opioid receptor
Benjamin J. Polacco, Braden T. Lobingier, Emily E. Blythe, Nohely Abreu, Prachi Khare, Matthew K. Howard, Alberto J. Gonzalez-Hernandez, Jiewei Xu, Qiongyu Li, Brandon Novy, Zun Zar Chi Naing, Brian K. Shoichet, Willow Coyote-Maestas, Joshua Levitz, Nevan J. Krogan, Mark Von Zastrow† & Ruth Hüttenhain†
Nature Chemical Biology,
2024
Probing ion channel functional architecture and domain recombination compatibility by massively parallel domain insertion profiling
Willow Coyote-Maestas, David Nedrud, Antonio Suma, Yungui He, Kenneth A Matreyek, Douglas M Fowler, Vincenzo Carnevale, Chad L Myers, Daniel Schmidt
Nature communications,
2021
A large‐scale survey of pairwise epistasis reveals a mechanism for evolutionary expansion and specialization of PDZ domains
David Nedrud, Willow Coyote‐Maestas, Daniel Schmidt
Protein Science,
2021
Targeted insertional mutagenesis libraries for deep domain insertion profiling
Willow Coyote-Maestas*, David Nedrud**, Steffan Okorafor, Yungui He, Daniel Schmidt
Nucleic Acids Research,
2019
Domain insertion permissibility-guided engineering of allostery in ion channels
Willow Coyote-Maestas, Yungui He, Chad L Myers, Daniel Schmidt
Nature communications,
2019
Two distinct pools of B12 analogs reveal community interdependencies in the ocean
Katherine R Heal, Wei Qin, Francois Ribalet, Anthony D Bertagnolli, Willow Coyote-Maestas, Laura R Hmelo, James W Moffett, Allan H Devol, E Virginia Armbrust, David A Stahl, Anitra E Ingalls
PNAS,
2017